Estimates the model by OLS based on the outcomes and model matrices provided in the outputs of lmpModelMatrix function and calculates the estimated effect matrices \(\hat{\mathbf{M}}_0, \hat{\mathbf{M}}_1, ...\hat{\mathbf{M}}_F\), ... and residual matrix \(\hat{\mathbf{E}}\). It calculates also the type III percentage of variance explained by each effect.
Arguments
- resLmpModelMatrix
A list of 5 elements from
lmpModelMatrix.- SS
Logical. If
FALSE, won't compute the percentage of variance for each effect.- contrastList
A list of contrasts for each parameter. If
NA, the function creates automatically the list by default.
Value
A list with the following elements:
lmpDataListThe initial object: a list with outcomes, design and formula.
modelMatrixA nxp model matrix specifically encoded for the ASCA-GLM method.
modelMatrixByEffectA list of F+1 model matrices for each effect.
effectsNamesUniqueA character vector with the F+1 names of the model effects, each repeated once.
effectsNamesAllA character vector with the p names of the model effects ordered and repeated as the column names of the model matrix.
effectMatricesA list of F+1 effect matrices for each model effect.
predictedvaluesThe nxm matrix of predicted outcome values.
residualsThe nxm matrix of model residuals.
parametersThe pxm matrix of the estimated parameters.
type3SSA vector with the type III sum of squares for each model effect (If SS = TRUE).
variationPercentagesA vector with the percentage of variance for each model effect (If SS = TRUE).
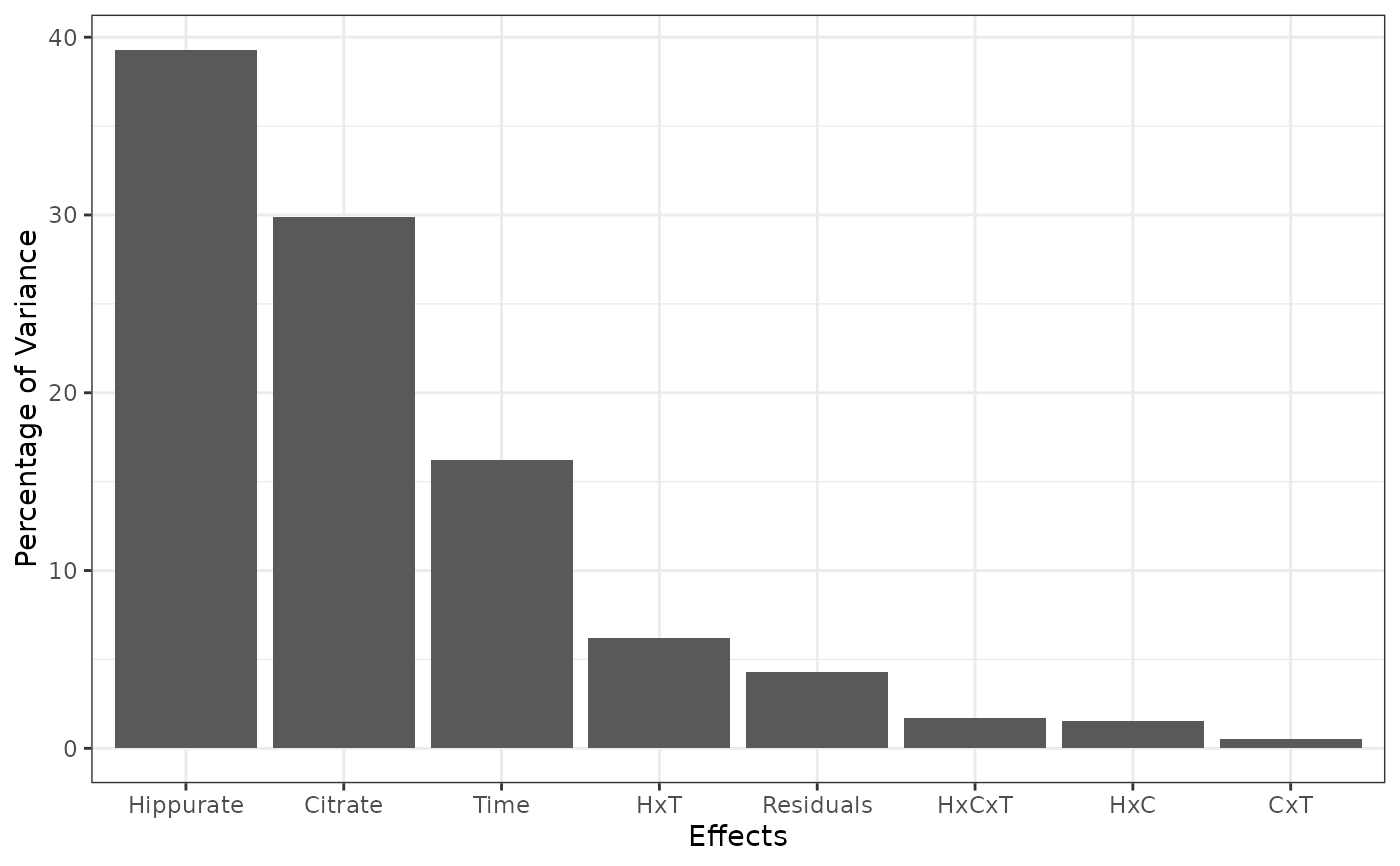
varPercentagesPlotA ggplot bar plot of the contributions of each model effect to the total variance (If SS = TRUE).
References
Thiel M.,Feraud B. and Govaerts B. (2017) ASCA+ and APCA+: Extensions of ASCA and APCA in the analysis of unbalanced multifactorial designs, Journal of Chemometrics
Examples
data("UCH")
resLmpModelMatrix <- lmpModelMatrix(UCH)
reslmpEffectMatrices <- lmpEffectMatrices(resLmpModelMatrix)
reslmpEffectMatrices$varPercentagesPlot